Moving ORD Practices into Cancer Care
About this Project
This project explores and tests ORD principles in the context of cancer care through a combination of engagement with relevant communities at the UZH as well as a technical demonstrator of a cutting edge data discovery technology. The cancer community at UZH is being mobilized through a dedicated workshop and topical meetings, to discuss challenges and opportunities of ORD in the oncology domain. A pilot installation of an established ORD approach (Beacon technology by the Global Alliance for Genomics and Health) is being implemented to test the sharing of cancer related -omics and associated data with a specific focus on the specific data access and security aspects presenting here.
Scientific Summary
The systematic and standardized sharing of routinely collected data is key for improving our abilities to optimize cancer treatment. Analysis of routine data allows to derive additional treatment evidence, complementing traditional evidence from Randomized Controlled Trials (RCTs) or related trial designs. Unlike RCTs, which require lengthy prospective data collections, are restricted to patients with specific inclusion criteria and are typically limited to a defined number of patients, data analysis on routinely collected data allows for the fast and immediate inspection of treatment choices across vast patient archives reflecting treatment outcomes across different patient and disease types. Since only around 5% of cancer patients world-wide are included in specific trial protocols, it is paramount to make routine data also available for research. Particularly, genomic characterization of patients’ cancers, using Next Generation Sequencing (NGS) or similar approaches, leads to an ever-increasing stratification of patients into small groups of individuals sharing specific genomic features that are essential for understanding their treatment outcomes. However, the traditional RCT setting does not allow for accruing enough patients from one of these groups and there is the necessity to resort to the analysis of larger sets of routinely collected patient information.
The Vision of this Project
The vision is as follows: as physicians decide on an optimal care plan, they consult all available treatment evidence including evidence from patients sharing equivalent genomic (or other -omics) features, with the assumption that past and successful treatment choices in these similar patients may be applied to currently treated patients too. To make this vision a reality, there is a need to massively collect, store and share comparable cancer data across institutions, as no single institution will harbor enough information about a group of patients sharing the same genomic features. The Open Research Data (ORD) paradigm could be ideally suited to support this vision.
Current Gaps and Challenges
However, there is little or only recent experience with ORD practices in clinical cancer care. On the contrary, the heterogeneous data collection and storage in Switzerland to date, as well as some regulations on human research and data protection, limit data sharing considerably. Recent initiatives, such as the Swiss Personalized Health Network (SPHN), which also supports research involving cancer data (Swiss Personalized Oncology National Data Stream), address some of the key ORD and FAIR requirements, including the standardization of data and making data interoperable. However, these initiatives offer only early solutions for massive, rapid and comprehensive data sharing across Swiss institutions with very limited current options for extended, federated data analyses or direct contributions to international data sharing efforts. Effective cancer data sharing should also involve a global alliance, as the stratification of cancer patients can only be addressed by world-wide data collection.
The Global Alliance for Genomics and Health
The Global Alliance for Genomics and Health (GA4GH) is a policy-framing and technical standards-setting organization which develops, implements and promotes ORD standards and directives in biomedical genomics and related areas. The University of Zurich and the Swiss Institute of Bioinformatics are among the founding institutions of GA4GH, and in 2019 the SPHN became accepted as a GA4GH "driver project" based on the explicit statement of support for GA4GH. Besides established formats such as BAM, CRAM or VCF, recent additions to the GA4GH portfolio includes the Beacon genomics discovery protocol, which is becoming the standard for discovery of genomics resources' data content and in facilitating the federated analysis of such data.
Our project utilizes the engagement with stakeholders from different areas relevant for data discovery and sharing approaches with relevance to the cancer care domain, including clinicians, genomics researchers as well as experts in legal aspects and information security, to explore avenues towards enabling the utilization of genomic and related data in scenarios following the main ORD “as open as possible, as protected as necessary” paradigm.
Also, implementation of emerging ORD international data standards such as Beacon will allow the utilization of data generated in national projects / data streams in an international context.

Challenges and Goals
Goal 1: Bring Stakeholders together and Raise Awareness
The goal of this project lies in the conceptual design of and build an open research database for oncology patient data matching, tailored to the local prerequisites and challenges of the Zurich biomedical environment. To this end, we explore (in line with but expanding beyond the previous and continuing work of the SPHN Swiss Personalized Oncology project) what legal, medical and technical framework is necessary for such an approach partially by mobilizing stakeholders to discuss the challenges and opportunities of ORD practices in cancer care. An additional aspect here is in spreading awareness of the importance of ORD aspects for local stakeholders and potential external interactions.
Read more...
A main tool is a workshop "ORD in cancer care" to discuss the political, ethical, legal and technical challenges in as perceived by participants representing different aspects such as clinical and research genomics as well as legal and ethical aspects. Here, he project leads (Profs. Krauthammer, Wicki, Baudis, Tag) can rely on their extensive network of colleagues for assembling a cadre of experts that are capable of discussing the different aspects of changing the data sharing culture in cancer. The output of the workshop is ideally a position / white paper on the topic.
Also, as part of the study a number of smaller meetings on subjects such as Beacon technology, open cancer data, cancer data models and others that are crucial building blocks of the ORD paradigm, partially by inviting domain specific experts but also through participation at the 2024 "Connect" meeting of the Global Alliance for Genomics and Health (GA4GH).
Goal 2: Develop Data Discovery Technology
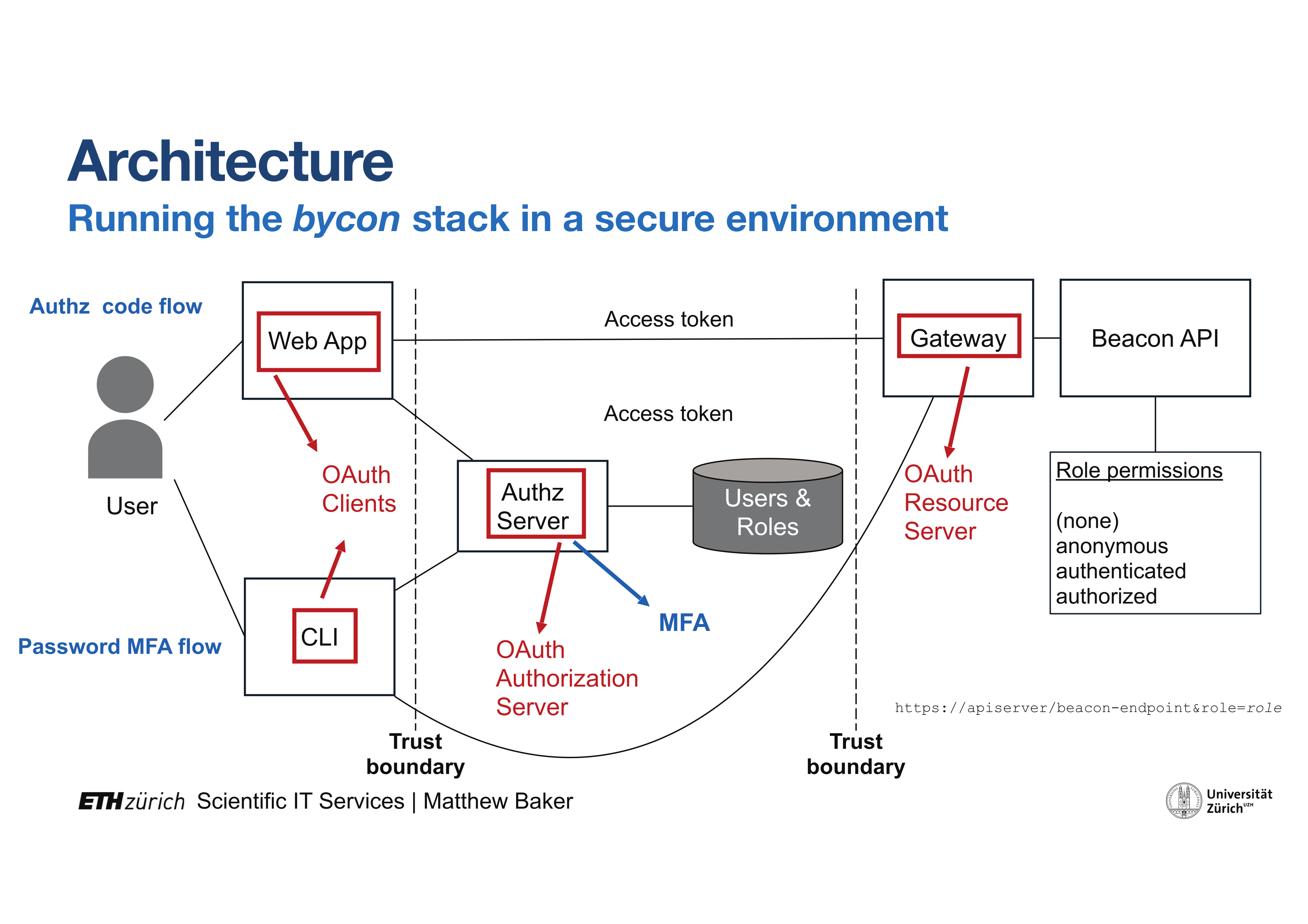
The second objective of the project was in the implementation-based evaluation of challenges from the establishment of a data discovery technology for cancer related genome-phenome data under the conditions relevant for oncology practitioners and researchers in the Zurich biomedical environment. Here, a reference implementation of the leading edge GA4GH Beacon protocol was selected for testing on a secure computer system (LeoMed). One main focus of the Beacon test scenario is in close collaboration with LeoMed Security engineers for a detailed risk assessment and development of a tailored authentication and authorization concept. Focus meetings are employed to evaluate practical hurdles in moving CCCZ data into the Beacon reference implementation.
Results and Output
- Workshop on ORD in cancer care (June 2024)
- White paper or similar workshop summary
- "Connect" meeting of GA4GH
- Topic meetings
- Installed Beacon platform on LeoMed secure computing platform, including development of an access and authorization model
Impact on Open Science Practices
We anticipate that the use of real-world data will increasingly refine and inform therapy decisions beyond current guidelines. In the long run, this may result in better outcomes and at the same time help to get a grasp on costs since the likelihood of applying ineffective and/or toxic therapies will decrease. ORD practices testing and explored in this grant will foster other precision oncology projects, by providing data to an expanding ecosystem of re-usable -omics and observational data.